Prediction of Molar Extinction Coefficients of Proteins and Peptides Using UV Absorption of the Constituent Amino Acids at 214 nm To Enable Quantitative Reverse Phase High-Performance Liquid Chromatography−Mass Spectrometry Analysis | Journal
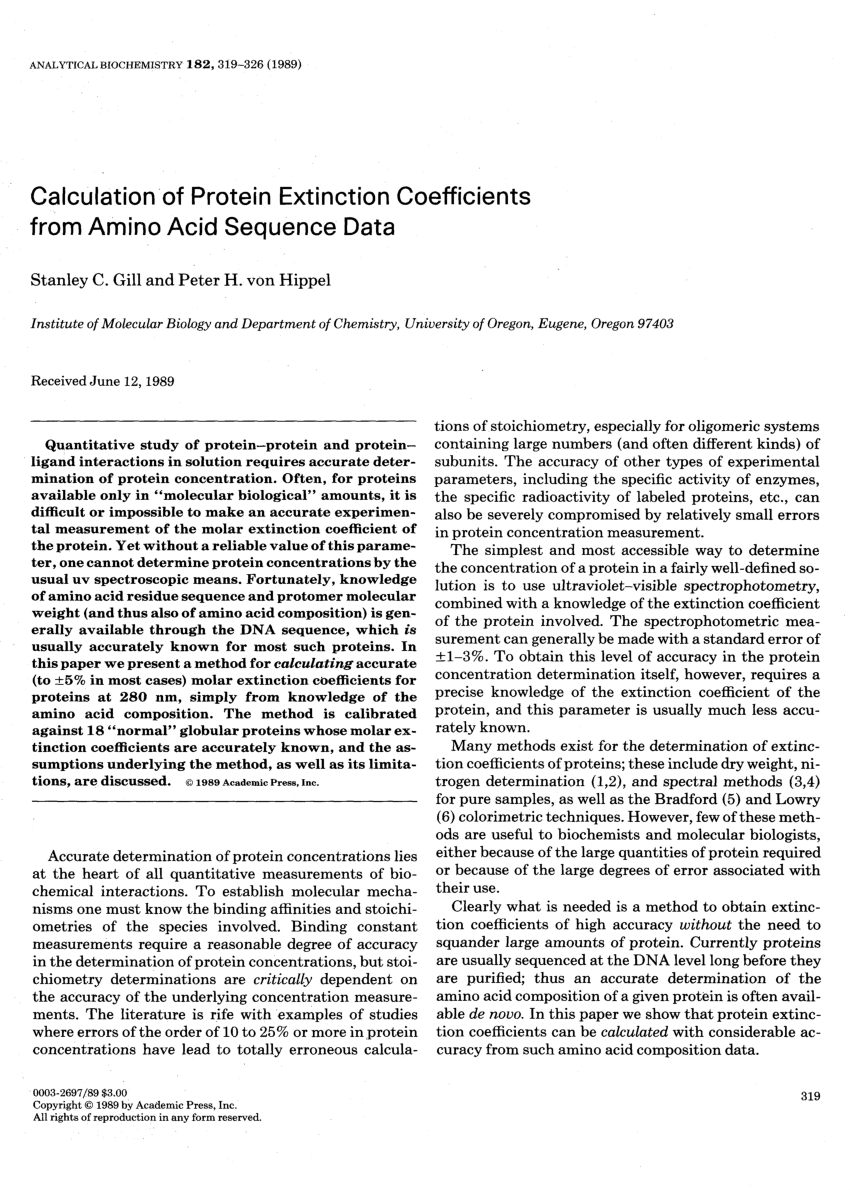
![PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/af80a5d2beee749cff2f5d98dc4a8f0dfdd48074/3-Table2-1.png)
PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar

Molecules | Free Full-Text | Evaluation of Peptide/Protein Self-Assembly and Aggregation by Spectroscopic Methods

A method to identify and quantify the complete peptide composition in protein hydrolysates - ScienceDirect

SOLVED: It is possible to estimate the molar extinction coefficient of a protein from knowledge of its amino acid composition, as shown from your experiences with ExPASY: From the molar extinction coefficient

Prediction of Molar Extinction Coefficients of Proteins and Peptides Using UV Absorption of the Constituent Amino Acids at 214 nm To Enable Quantitative Reverse Phase High-Performance Liquid Chromatography−Mass Spectrometry Analysis | Journal
![PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/af80a5d2beee749cff2f5d98dc4a8f0dfdd48074/2-Table1-1.png)
PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar
![PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/af80a5d2beee749cff2f5d98dc4a8f0dfdd48074/4-Table3-1.png)
PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar

Photometric Quantification of Proteins in Aqueous Solutions via UV-Vis Spectroscopy - Eppendorf Handling Solutions